Thrust 3: Adaptive Cellular Communication (Woo lead).
The research theme is cellular communication as the key regulator in biological processes that involve assemblies of cells and their extracellular environment (development, wound healing, cancer progression). Cells move by adhering to and exerting mechanical forces on adjacent cells and the extracellular matrix and, in turn, respond to the feedback from such communication. Phase II will examine the impact of such cell-cell and cell-matrix mechanical interactions on collective cell motility, patterning and the emergence of function, combining experimental and modeling approaches, and organized in two broad areas.
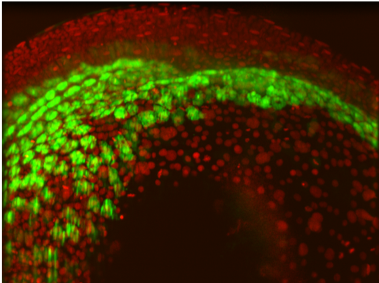
3.1. Developing Tissue. In Phase I we analyzed combinatorial factors in development [30] and developed a 2D experimental and computational model of differentiating vascular endothelial and smooth muscle cells. For Phase II we will incorporate cell-cell signaling and mechanical interactions leading to a 3D agent-based experiment-simulation model. As an experimental system, we will use co-cultures of endothelial and mural cells that assemble into perfusable microvascular structures to explore the signaling (cell-cell and cellmatrix) required for directing the assembly of vascular lumens with controlled dimensions [31-33]. New Phase II projects aim at studying mesenchymal to epithelial transitions in the zebrafish endoderm using fluorescence microscopy, force spectroscopy and computational modeling [34,35]; or how alcohol rearranges neural synapses, using super-resolution microscopy and mass spectrometry to identify the signaling transducers in the primary cilia of neural stem cells.
3.2. Bacterial Community Motility. Communities of motile bacteria exist in many environments ranging from the soil to human tissue [36,37]. These communities exhibit emergent spatial structuring and dynamic 3 patterns that are not well understood. In Phase I, we developed an approach to follow individual bacteria over long times through a micropillar array (a structured environment). We showed that, in non-adaptive mutant strains, minor variations in cell size led to large differences in transport. In Phase II, we propose to follow wild type bacteria in specially developed strain-free Stokes “wind tunnels” to understand the cell’s adaptive response to surface mechano-sensing or interactions with other bacteria in communities. A new Phase II project will explore the impact of bacterial swarming [38] on the phylogenetic spatial structure of a bacterial population [39], with implications for the evolution of highly antibiotic-resistant strains.
Crosscutting Thrusts 1 and 3.
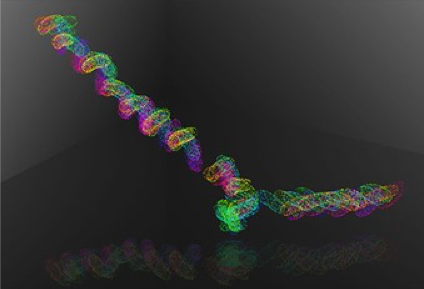
In response to emerging synergies in gene expression between thrusts 1 and 3, we propose a new Phase II project to investigate the control of phenotype switching between the white and opaque cell states of Candida albicans. This transcriptional regulatory circuit closely mirrors the equivalent circuitry that maintains pluripotency in mammalian stem cells [40]. We will focus on three critical transcription factors: Wor1, the master regulator of the opaque state; Efg1, a primary stabilizer of the white cell state and repressor of Wor1; and Wor3, a factor important for temperature-induced state switching. The three factors have disordered (morphing) regions that drive phase separation and recruit other partners into these condensates [40]. We will explore how Efg1 (alone or with cofactors) binding to DNA changes as conditions are driven toward microphase separation using single-molecule microscopy, use high-throughput SELEX-seq to quantify Efg1 DNA binding profiles, and high-throughput genomics to map the in vivo binding sites, chromatin accessibility, and the effects of knocking out TFs and specific binding sites edited with CRISPR-Cas9.